CellSegmentation
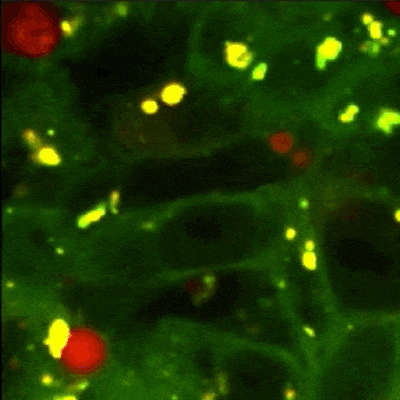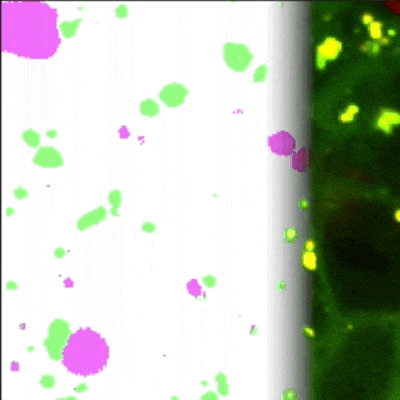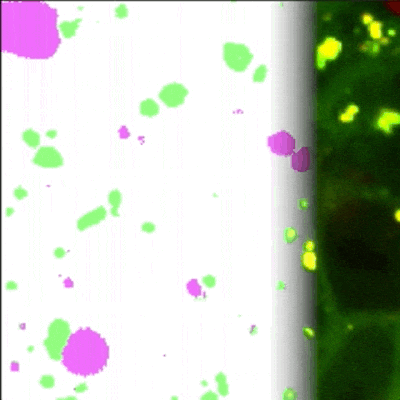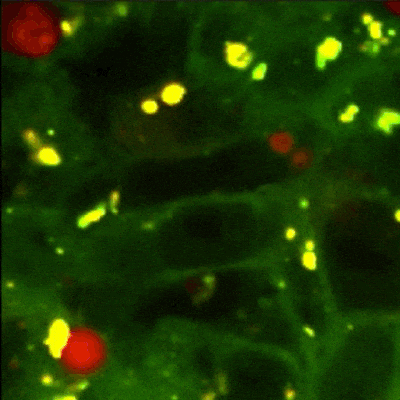
MATLAB GUI tool for image segmentation of autophagy events detected with Rosella pH-sensitive biosensor in high-throughput high-content microscopy images.
CellSegmentation GUI for MATLAB
Welcome to the instructions to install CellSegmentation GUI for MATLAB
Cite as
Paper coming soon :-)
Preprint available at Research Square.
Beatriz García Santa Cruz, Jan Sölter, Gemma Gomez Giro et al. Generalising from Conventional Pipelines: A Case Study in Deep Learning-Based for High-Throughput Screening, 29 October 2021, PREPRINT (Version 1) available at Research Square. https://doi.org/10.21203/rs.3.rs-991404/v1
Description
The study of complex diseases relies on large amounts of data to build models toward precision medicine. Such data acquisition is feasible in the context of high-throughput screening, in which the quality of the results relies on the accuracy of the image analysis. Although state-of-the-art solutions for image segmentation employ deep learning approaches, the high cost of manually generating ground truth labels for model training hampers the day-to-day application in experimental laboratories.
Alternatively, traditional computer vision-based solutions do not need expensive labels for their implementation. Our work combines both approaches by training a deep learning network using weak training labels automatically generated with conventional computer vision methods. Our network surpasses the conventional segmentation quality by generalising beyond noisy labels, providing a 25% increase of mean intersection over union, and simultaneously reducing the development and inference times.
Our solution was embedded into an easy-to-use graphical user interface that allows researchers to assess the predictions and correct potential inaccuracies with minimal human input. To demonstrate the feasibility of training a deep learning solution on a large dataset of noisy labels automatically generated by a conventional pipeline, we compared our solution against the common approach of training a model from a small manually curated dataset by several experts. Our work suggests that humans perform better in context interpretation, such as error assessment, while computers outperform in pixel-by-pixel fine segmentation. Such pipelines are illustrated with a case study on image segmentation for autophagy events.
This work aims for better translation of new technologies to real-world settings in microscopy-image analysis.
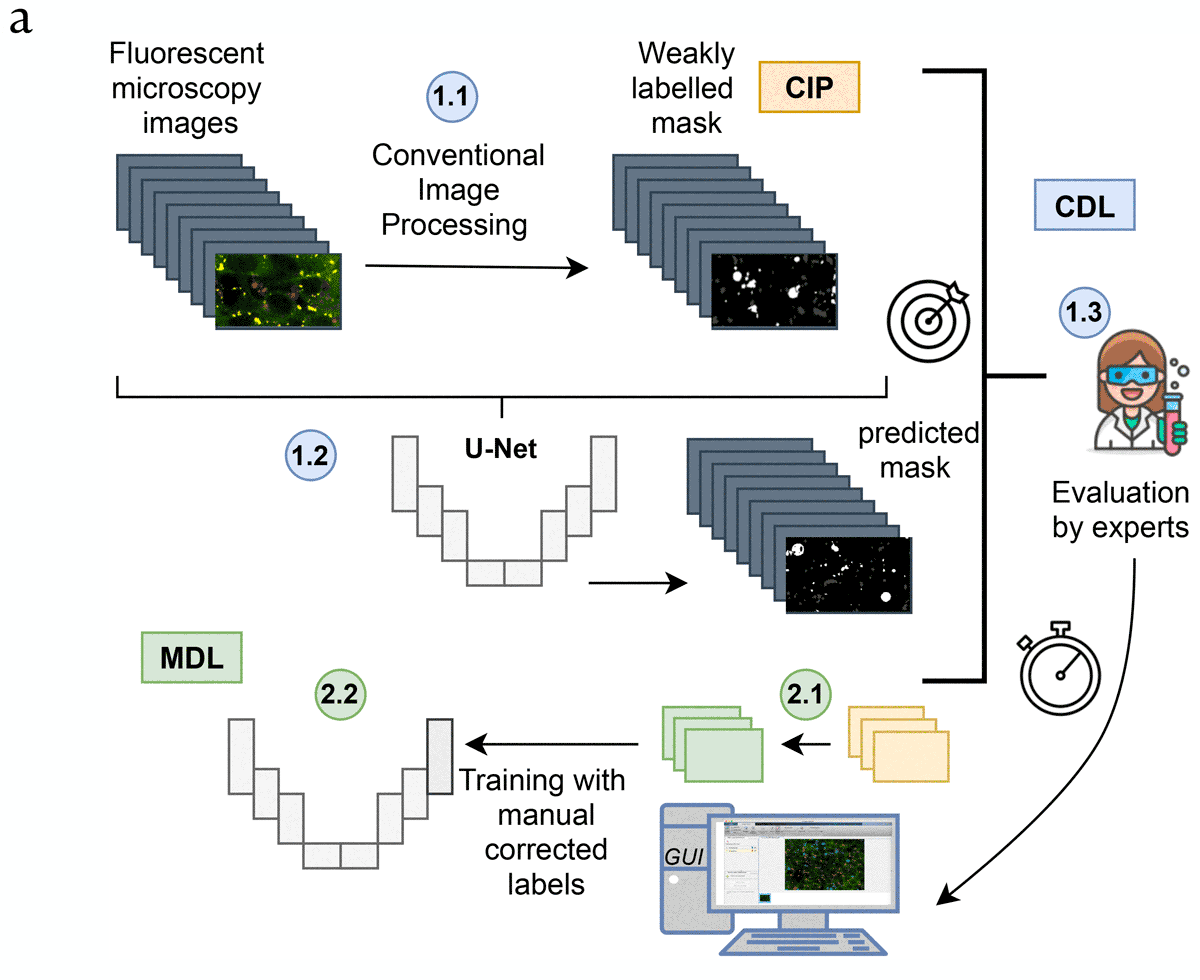
Panel A: General workflow of the proposed pipeline, part 1: (In blue, CIP-based DL: CDL). [1.1] First a weakly labelled dataset is created using conventional imaging processing (CIP). [1.2] After that, a U-net like architecture is trained and [1.3] the accuracy of the evaluated. Including an integration of the trained network in an intuitive tool for biologists that allows easy correction.
Proposed pipeline, part 2: (In green, for Manually based DL: MDL). [2.1] Manual corrected masks are easily generated using the GUI, which is employed to train a U-net from scratch [2.2].
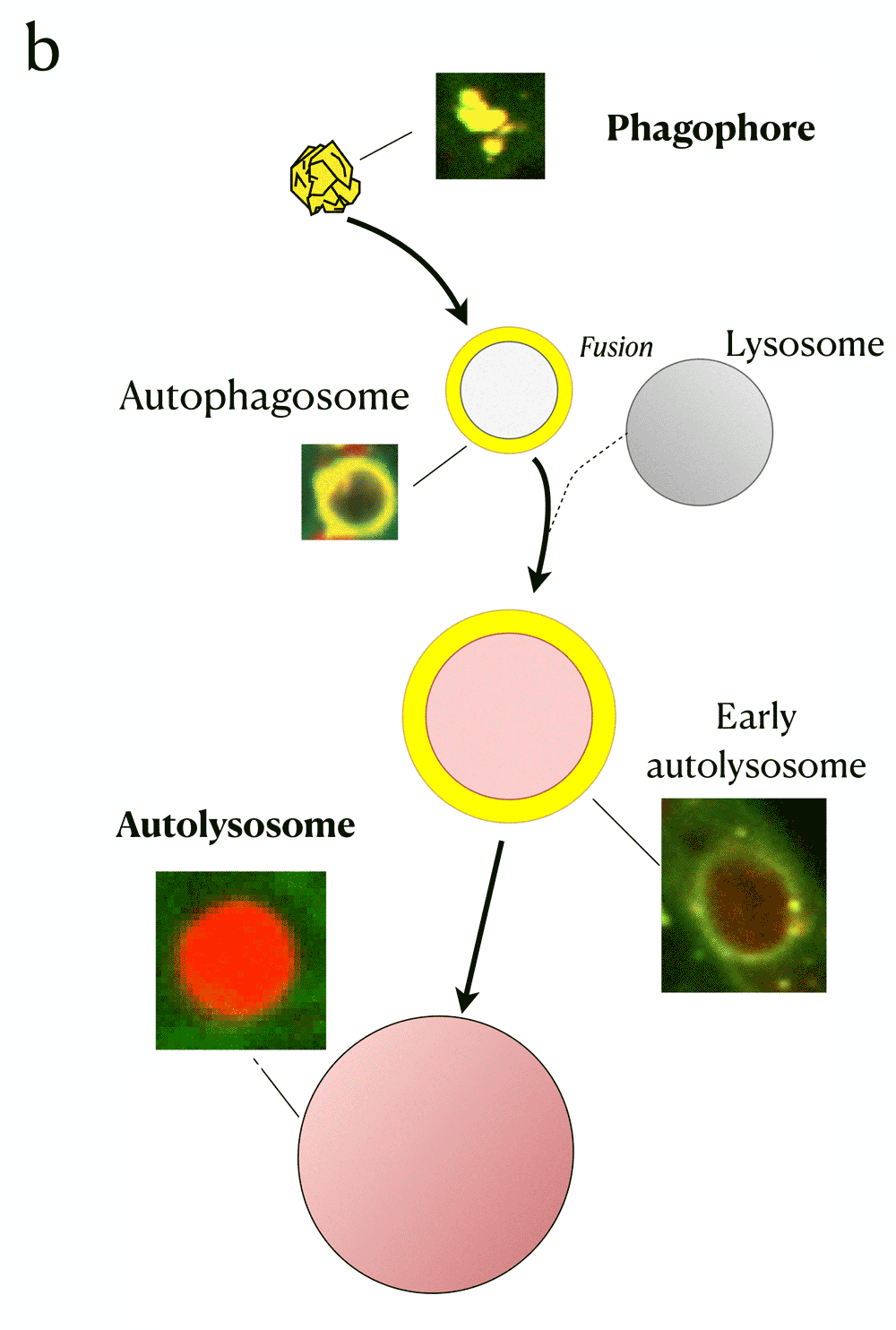
Panel B: The biological process of autophagy and its detection with Rosella biosensor. The four main phases are: The initial state - Phagophore, intermediate states~- Autophagosome and early autolysosome, and final state - Autolysosome. The fusion with the lysosome during the autophagy process yields a pH decrease which induces a change of colour in the fluorescent microscopy image.
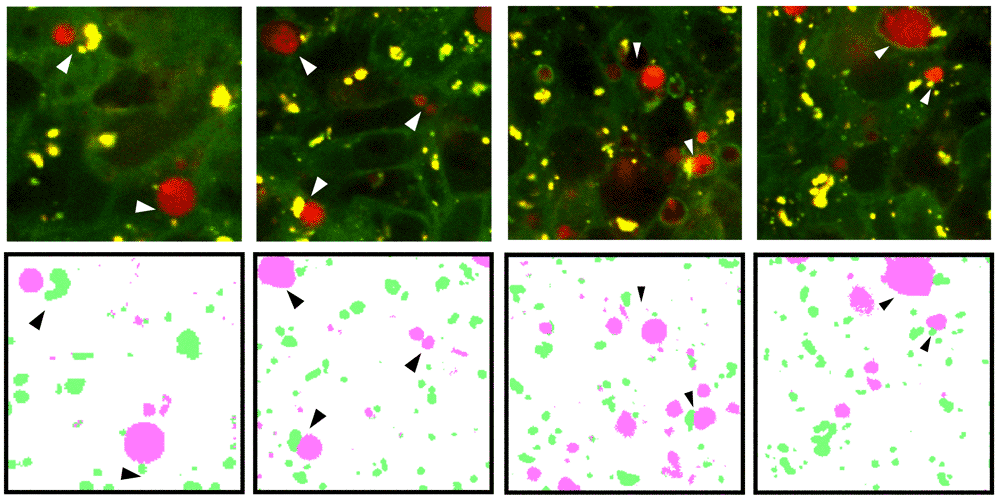
Instructions
To install and use the GUI in MATLAB:
- Use Matlab 2018 or later.
- Install the following toolboxes: Image Labeler, Image Analysis, Deep Learning.
- Download the folder integration from this repository.
- Open Image Labeler toolbox.
- Load the abels from
Labels_manual_curation.matfile. - Add images from folder.
- Add the new algorithm selecting the file
AutophagyDeepLearningSegmentation.mlocated inintegration/+vision/+labeler/folder. - Run the algorithm clicking on Automate and Run.
- Follow the video for more details on how to improve the segmentation.
Video demo
The following video depicts how to add the algorithm to the image labeler GUI in MATLAB and use it for Cell Segmentation.
Acknowledgments
- National Department of Neurosurgery, Centre Hospitalier de Luxembourg.
- Interventional Neuroscience Group, Luxembourg Center for Systems Biomedicine, University of Luxembourg.
- Developmental and Cellular Biology, Luxembourg Center for Systems Biomedicine, University of Luxembourg.
- Systems Control Group, Luxembourg Centere for Systems Biomedicine, University of Luxembourg.